Difference between revisions of "Research"
(→Agent Based Model of the infection spread within a small population of Guinea pigs, dependent on temperature and humidity conditions of the surroundings) |
(→Alignment-free genome sequence analysis) |
||
| Line 44: | Line 44: | ||
==== Alignment-free genome sequence analysis ==== | ==== Alignment-free genome sequence analysis ==== | ||
| + | |||
| + | [[Image:occurence_probability.gif |frameless|300px|right]] | ||
Synonymous codons do not occur at equal frequencies. Codon usage and codon bias have been extensively studied. However, the sequential order in which synonymous codons appear within a gene has not been studied until now. Here we describe an in silico method, which is the first attempt to tackle this problem: to what extent this sequential order is unique, and to what extent the succession of synonymous codons is important. This method, which we called Intragenic, Stochastic Synonymous Codon Occurrence Replacement (ISSCOR), generates, by a Monte Carlo approach, a set of genes which code for the same amino acid sequence, and display the same codon usage, but have random permutations of the synonymous codons, and therefore different sequential codon orders from the original gene. We analyze the complete genome of the bacterium Helicobacter pylori (containing 1574 protein coding genes), and show by various, alignment-free computational methods (e.g., frequency distribution of codon-pairs, as well as that of nucleotide bigrams in codon-pairs), that: (i) not only the succession of adjacent synonymous codons is far from random, but also, which is totally unexpected, the occurrences of non-adjacent synonymous codon-pairs are highly constrained, at strikingly long distances of dozens of nucleotides; (ii) the statistical deviations from the random synonymous codon order are overwhelming; and (iii) the pattern of nucleotide bigrams in codon-pairs can be used in a novel way for characterizing and comparing genes and genomes. Our results demonstrate that the sequential order of synonymous codons within a gene must be under a strong selective pressure, which is superimposed on the classical codon usage. This new dimension can be measured by the ISSCOR method, which is simple, robust, and should be useful for comparative and functional genomics. | Synonymous codons do not occur at equal frequencies. Codon usage and codon bias have been extensively studied. However, the sequential order in which synonymous codons appear within a gene has not been studied until now. Here we describe an in silico method, which is the first attempt to tackle this problem: to what extent this sequential order is unique, and to what extent the succession of synonymous codons is important. This method, which we called Intragenic, Stochastic Synonymous Codon Occurrence Replacement (ISSCOR), generates, by a Monte Carlo approach, a set of genes which code for the same amino acid sequence, and display the same codon usage, but have random permutations of the synonymous codons, and therefore different sequential codon orders from the original gene. We analyze the complete genome of the bacterium Helicobacter pylori (containing 1574 protein coding genes), and show by various, alignment-free computational methods (e.g., frequency distribution of codon-pairs, as well as that of nucleotide bigrams in codon-pairs), that: (i) not only the succession of adjacent synonymous codons is far from random, but also, which is totally unexpected, the occurrences of non-adjacent synonymous codon-pairs are highly constrained, at strikingly long distances of dozens of nucleotides; (ii) the statistical deviations from the random synonymous codon order are overwhelming; and (iii) the pattern of nucleotide bigrams in codon-pairs can be used in a novel way for characterizing and comparing genes and genomes. Our results demonstrate that the sequential order of synonymous codons within a gene must be under a strong selective pressure, which is superimposed on the classical codon usage. This new dimension can be measured by the ISSCOR method, which is simple, robust, and should be useful for comparative and functional genomics. | ||
<!--[[Genetic:isscor | '''More..''']]--> | <!--[[Genetic:isscor | '''More..''']]--> | ||
</blockquote> | </blockquote> | ||
Revision as of 12:51, 19 May 2009
| Rivers | ||||
|---|---|---|---|---|
| Research | Software | Publications | People | |
<br\>
EPIDEMIC SIMULATIONS
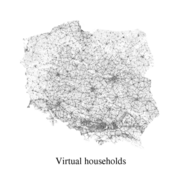
A virtual Polish society
Virtual society exists only in the computer memory. However, in our model, it represents and reproduces the real, Polish, society. Virtual society consists of individual (distinguishable) agents, each assigned to certain, geo-referenced, household. Further, household inhabitants, depending on their age should be either retired, employed or going to school (kindergarten, primary school, secondary school, college, university). All these basic relations should be reflected in the virtual model. This virtual society was based on accessible data. Depending on the data available and on the particular infrastructure element that was to be incorporated into the virtual society, we developed a set of methods. More...
Agent Based Model of the infection spread within a small population of Guinea pigs, dependent on temperature and humidity conditions of the surroundings
The influence that atmospheric conditions might have on the efficiency of the spread of influenza virus is important for epidemiological and evolutionary research. However, it has not been satisfactorily recognized and quantified so far. Here we provide a statistical model of influenza transmission between individuals. It has been derived from the results of recent experiments, which involved infecting guinea pigs with influenza at various temperatures and relative air humidity levels. The wide range of transmission rates in those experiments reflects the ensemble-independent phenomena. The correlation between most of our simulations and the experimental results is satisfactory. For several different conditions, we obtained transmissibility values which seem to be sufficiently accurate to provide partial input for an intended large-scale epidemiological study in the near future. More..
GENETIC STUDIES
Alignment-free genome sequence analysis
Synonymous codons do not occur at equal frequencies. Codon usage and codon bias have been extensively studied. However, the sequential order in which synonymous codons appear within a gene has not been studied until now. Here we describe an in silico method, which is the first attempt to tackle this problem: to what extent this sequential order is unique, and to what extent the succession of synonymous codons is important. This method, which we called Intragenic, Stochastic Synonymous Codon Occurrence Replacement (ISSCOR), generates, by a Monte Carlo approach, a set of genes which code for the same amino acid sequence, and display the same codon usage, but have random permutations of the synonymous codons, and therefore different sequential codon orders from the original gene. We analyze the complete genome of the bacterium Helicobacter pylori (containing 1574 protein coding genes), and show by various, alignment-free computational methods (e.g., frequency distribution of codon-pairs, as well as that of nucleotide bigrams in codon-pairs), that: (i) not only the succession of adjacent synonymous codons is far from random, but also, which is totally unexpected, the occurrences of non-adjacent synonymous codon-pairs are highly constrained, at strikingly long distances of dozens of nucleotides; (ii) the statistical deviations from the random synonymous codon order are overwhelming; and (iii) the pattern of nucleotide bigrams in codon-pairs can be used in a novel way for characterizing and comparing genes and genomes. Our results demonstrate that the sequential order of synonymous codons within a gene must be under a strong selective pressure, which is superimposed on the classical codon usage. This new dimension can be measured by the ISSCOR method, which is simple, robust, and should be useful for comparative and functional genomics.